The Human Genome Project, the first successful effort to sequence the entire human genome, was completed in 2003 after 13 years of relentless work by researchers. The technology that was used for this project, called Sanger sequencing, was able to cover about 92% of the DNA, and it was the only available option at the time. Today, with significant advancements in the field of genomics, researchers and consumers have more DNA sequencing technologies to choose from.
While Sanger is still quite popular, Illumina is another leading sequencing provider that has gained traction over the years. To give you context, Illumina’s next-generation sequencing (NGS) was specifically created to address Sanger’s limitations, potentially making it a more modern and efficient sequencing method.
You can learn more about the differences between these two methods/technologies in our thorough Illumina sequencing vs. Sanger sequencing breakdown. We’ll explain how they work, as well as cover aspects like cost, accuracy, and applications, especially for deep genetic health assessments.
Sanger: The gold standard of sequencing technology
Sanger sequencing, also called the chain termination method and dideoxy sequencing, was invented in 1977 by Frederick Sanger. Besides being the first, it was also the most widely used sequencing technology for about 30 years.
The Sanger method played a crucial role in achieving the DNA sequencing outcomes in the Human Genome Project. The only major hiccup — the technology could only read one small DNA fragment at a time, which made assembling the genome a slow and laborious process. This time sink was one of the reasons why the project took over a decade to complete.
Sanger sequencing is still used extensively today because it’s over 99% accurate. Usually, it’s the preferred method when it comes to supporting small projects that aim to identify structural DNA variants (large insertions, deletions, or rearrangements), where Sanger proves to be highly effective. However, the growing need for larger-scale and consumer-grade DNA testing has led to the development of more efficient sequencing technologies (such as Illumina).

Source: Georgia Institute of Technology
Illumina: Fast, affordable next-generation sequencing (NGS)
NGS, the sequencing technology synonymous with Illumina, was introduced in 2000 by Solexa, a company that was later acquired by Illumina. NGS is a high-efficiency sequencing method that involves rapid sequencing of the whole genome or large amounts of DNA data parallelly. The technology has evolved at an impressive rate by increasing its data output per run by more than double each year, even surpassing Moore’s law.
Illumina’s NGS is among the most prevalent technologies today because of its competitive processing efficiency. Also, the company is one of the largest distributors of sequencing instruments, which are used by numerous DNA testing services, including:
By sharing its NGS technology with other organizations, Illumina has brought about faster turnaround and lower costs for larger DNA projects. The platform has especially made whole-genome sequencing (the most advanced sequencing level for rare and deep genetic insights) more accessible and affordable to consumers.
Bonus read: Find out how Illumina sequencing compares to PacBio’s third-generation sequencing method.

Source: University of Georgia
Sanger sequencing vs. Illumina sequencing: 6 key differentiators
Although similar in their core processes and goals, Sanger and Illumina sequencing have many distinct characteristics that make each suitable for different applications. Here’s a brief overview of the technologies to help set the stage for a more comprehensive comparison:
Now, let's better understand the two technologies, focusing on the following key aspects:
Sequencing process and read length
Sequencing volume and throughput
Accuracy and sensitivity
Potential costs
Reputation and perceived applications
1. Sequencing process and read length
The sequencing process for Sanger and Illumina is similar in its core. Both methods use the enzyme DNA polymerase to add nucleotides one by one and thus form a strand. They also use fluorescent tags to mark the nucleotides so the machine can later identify them and determine the DNA sequence.
Still, the two processing technologies differ in almost every other aspect, including read lengths. Read lengths refer to the number of DNA base pairs that can be sequenced at a time, which can impact the accuracy of analyses. Typically:
Short-read sequencing analyzes small DNA fragments at a time with high accuracy and low error margins.
Long-read sequencing analyzes relatively longer fragments of DNA at a time and can sequence repetitive regions or larger structural genetic variants better.
Illumina primarily uses short-read sequencing, which is the most commonly used method in the industry today. It’s worth noting that the company also launched a product that enables both short and long-read sequencing technology in 2023. Because of this, Illumina can reap the benefits of both methods, listed in the table below:
Sanger has long-read capabilities, but its read length falls somewhere in the middle when compared to Illumina. Sanger offers a longer read length than Illumina’s short-reads but a lower coverage than the latter’s new long-read sequencing technology. Still, Sanger remains a highly used and valued method due to its consistently high accuracy and familiar workflow.
2. Sequencing volume and throughput
The key difference between Sanger and Illumina sequencing is in the amount of DNA data they can process in a single run. While Sanger can analyze only one fragment at a time, Illumina can read millions of DNA fragments in parallel, analyzing up to 1,000 times more genes per assay than Sanger.
Also, Illumina’s newly released long-read sequencing technology is expected to deliver an unprecedented level of throughput and accuracy, more than other similar products on the market.
Because of the higher volume and efficiency, Illumina, and especially its short-read sequencing technology, are more suitable for projects that entail high sample volumes.
Source: Illumina
Bonus read: We’ve evaluated today’s most popular DNA testing services. If interested, you can explore our reviews below:
3. Accuracy and sensitivity
Although it’s not as efficient as Illumina, Sanger sequencing is known for producing highly accurate results without the need for complex data analysis. Sanger sequencing, with an accuracy rate of 99.99%, is less error-prone and thus more likely to detect:
Single nucleotide polymorphisms, or SNPs, small variations at a single DNA position that occur among individuals
Variants that may not be present in every DNA copy of the sample
Illumina sequencing is also highly accurate, with an error rate of <1% (99.9+% accuracy) for detecting SNPs. Sanger is commonly used to fill in gaps and verify the findings from Illumina’s NGS method.
Still, because of its sequencing depth, Illumina outperforms Sanger in terms of sensitivity by potentially detecting variants present in only 1% of the DNA molecules. Illumina is also superior when it comes to discovery power and can detect a greater number of novel variants by deep sequencing, i.e., sequencing the same region many times.
4. Potential costs
The cost of sequencing with both methods varies greatly depending on many factors, such as the number of samples and the sequencing coverage (i.e., the entire genome that offers complete results or a few select genes).
In general, when it comes to high-volume projects, Illumina is more economical than Sanger because it’s more efficient and can analyze millions of fragments at the same time.
Illumina has even managed to reduce the cost of advanced whole-genome sequencing over the years. By working with DNA testing platforms such as Nucleus, Illumina has made affordable and clinical-grade genetic testing more accessible to people. Curious? You can order Nucleus whole-genome kits for only [PRICE.KIT_SOLO.ONE] and access deep health insights about your physical and mental health, your genetic traits and variants, IQ scores, and much more!
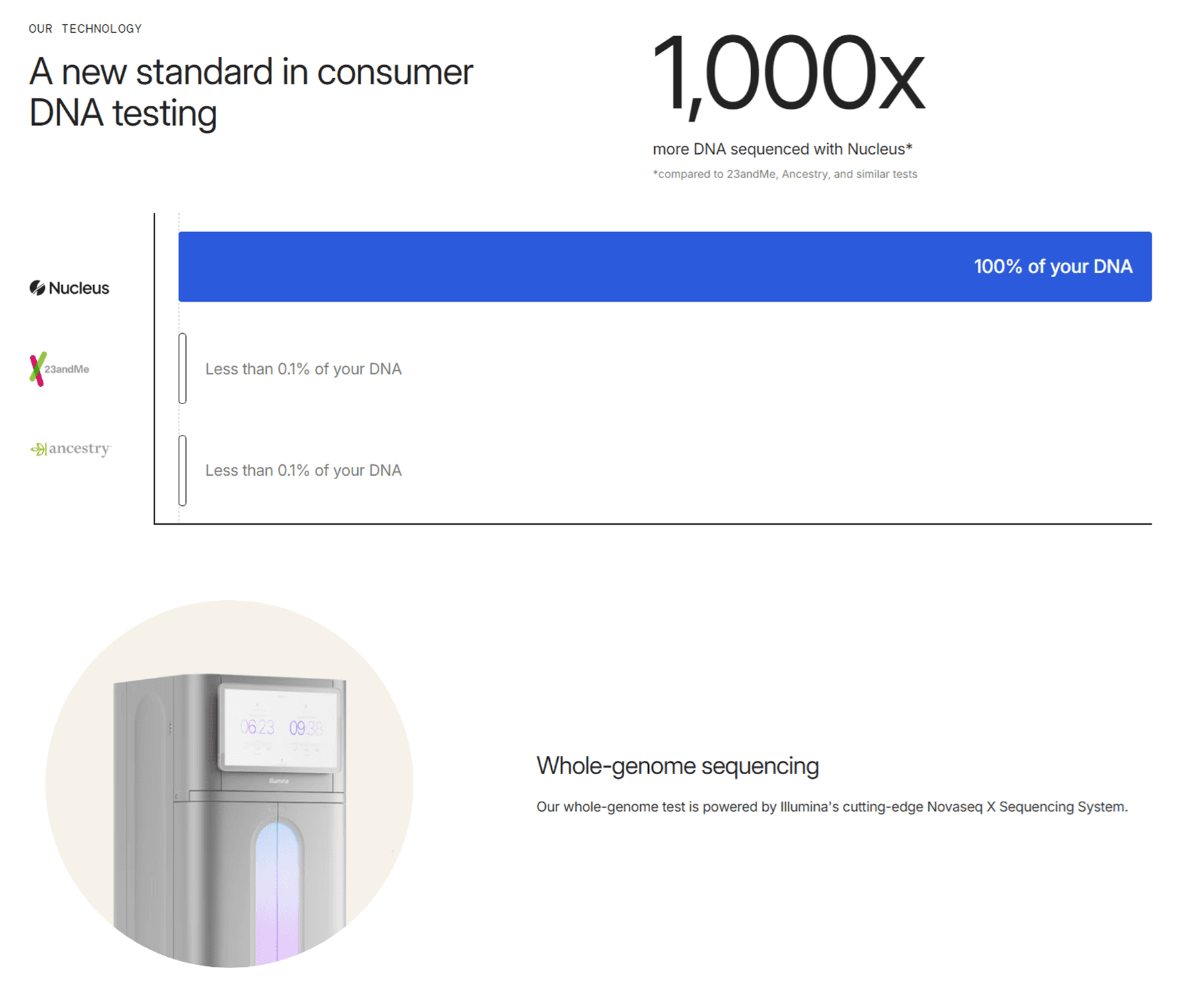
5. Reputation and perceived applications
Illumina and Sanger are both highly reputable and trustworthy sequencing methods, dominating the field of genomics for decades.
Sanger is known for its precision and reliability in small-scale applications or for more focused genetic studies, whereas Illumina brought us more efficient NGS, exome sequencing, and whole-genome sequencing that can be executed on larger scales. Illumina is usually the preferred method when large population samples are analyzed, for example, for cancer research, population studies, and understanding complex genetic diseases.
That said, Illumina as a company has consistently demonstrated commitment to innovation over the last few years, introducing new products that push the boundaries of genomic research and aim to make it even more scalable and accessible. Many genetics-oriented platforms find it to be the best sequencing company in the market.
Sanger vs. Illumina: Final thoughts
Both Sanger and Illumina sequencing have their unique strengths and applications, making both valuable depending on your testing goals.
Still, Illumina’s contribution to higher throughput and more accessible personal genomics space is particularly exciting. Illumina’s advanced technology now allows researchers to analyze nearly an entire genome and produce reports in under 30 hours.
This method, called whole-genome sequencing, is among the most powerful applications of sequencing, providing extensive and accurate insights into the person’s genetic health profile — some of which can be potentially life-saving.
One company that leverages Illumina’s whole-genome sequencing technology to make health-focused genetic information widely available is Nucleus. With its clinical-grade genetic test, Nucleus gets you 99.9% accurate insights on your disease risks and predispositions. As a result, you can exercise greater control over your health decisions and make genetics-informed lifestyle choices.
Start complete genetic self-discovery with Nucleus
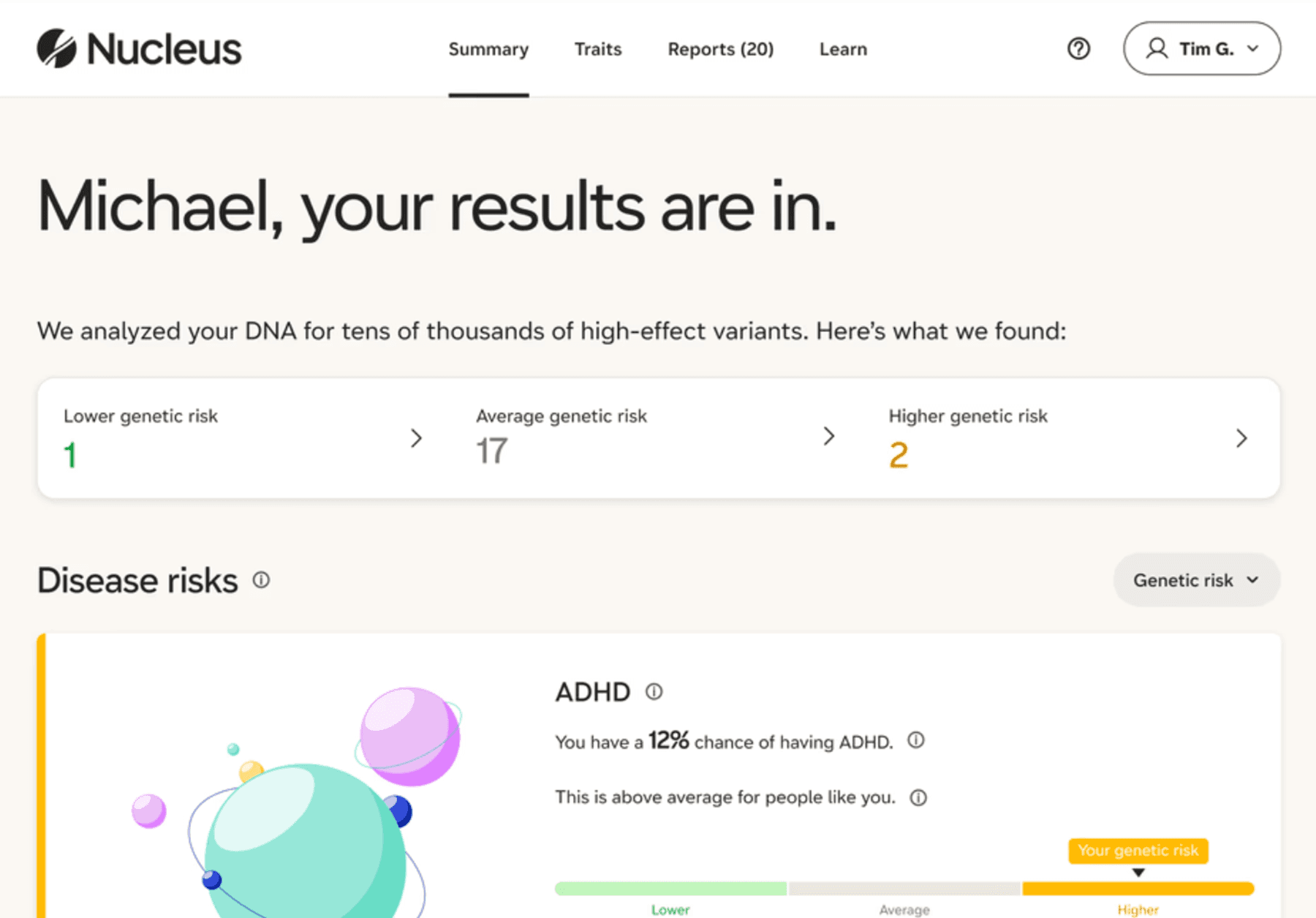
Nucleus is a DNA platform offering all-in-one health insights with its whole-genome DNA test kit. Once you provide a sample, Nucleus will analyze nearly all of your 6 billion DNA bases to provide a deep health assessment, targeting both variants with subtle impact and rare, high-effect disease-causing variants that other tests miss.
You can expect to learn about your risk for over 170 diseases and conditions, including common and rare ones, across categories such as:
Heart diseases
Neurological conditions
Food and diet
Cancers
Mental health

Source: Nucleus
Besides revealing how your genes predispose you to different conditions, Nucleus will consider your non-genetic factors like age and lifestyle choices to calculate personalized absolute risk scores. You can also find out if any hereditary conditions run in your family and get an analysis of your genetic traits, such as muscle strength, allergies, and intelligence scores.
Although Nucleus is a physician-approved test, it provides highly interpretive reports with clear language and illustrations. If you want actionable medical advice based on your results, Nucleus can arrange a session with a board-certified genetic counselor through its partnership with SteadyMD.
Nucleus is fully regulated and follows the highest industry standards to ensure quality, accuracy, and privacy. Besides being HIPAA-compliant, Nucleus works with CAP, CLIA-certified U.S. labs and sequences your DNA in the U.S.
Get your whole genome analysis with Nucleus
If you want to test your whole genome analyzed with Nucleus, you can:
Provide your personal information
Order the kit (HSA/FSA eligible)
Once you swab your cheeks and mail back the samples, you'll be able to view your reports online in 6–8 weeks.
You’ll get all available Nucleus reports at a single price point of [PRICE.KIT_SOLO.ONE]. The platform also updates reports for its members based on research advancements and the user’s lifestyle changes.
Source: Nucleus
You may also like…
We have numerous other DNA test reviews you can check out:
Featured image source: geralt












